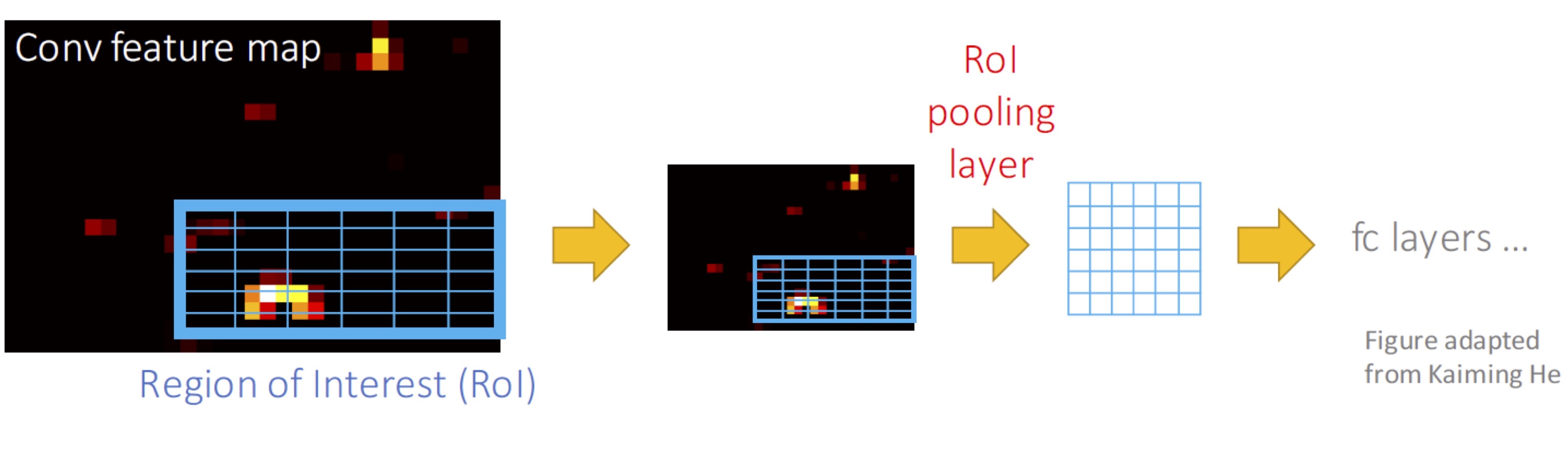
Umap Fast In R. The algorithm is founded on three assumptions about the data. Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. UMAP is a non linear dimensionality reduction algorithm in the same family as t-SNE. In the first phase of UMAP a weighted k nearest neighbour graph is computed, in the second a low dimensionality layout of this is then calculated. Runs the Uniform Manifold Approximation and Projection (UMAP) dimensional reduction technique. It's an independent implementation of UMAP (although it relied strongly on me looking at the code of the "real" Python UMAP package during its development). The umap R package also has its own independent implementation of UMAP (the default) or can use the Python UMAP if it's available to your R environment. UMAP is non-linear dimension reduction technique and often used for visualizing high-dimensional datasets. t-SNE and UMAP projections in R This page presents various ways to visualize two popular dimensionality reduction techniques, namely the t-distributed stochastic neighbor embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP).

Umap Fast In R. Its details are described by McInnes, Healy, and Melville and its official implementation is available through a python package umap-learn. Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. UMAP was created by Leland McInnes and John Healy ( github, arxiv ). This package provides an interface for two implementations. Just brute force will be a lot better. Umap Fast In R.
It's an independent implementation of UMAP (although it relied strongly on me looking at the code of the "real" Python UMAP package during its development).
UMAP was created by Leland McInnes and John Healy ( github, arxiv ).
Umap Fast In R. So what does UMAP bring to the table? The Riemannian metric is locally constant (or can be approximated as such); The manifold. Its details are described by McInnes, Healy, and Melville and its official implementation is available through a python package umap-learn. UMAP is a novel machine learning method for visualizing high-dimensional datasets. Here we will take a brief look at the performance characterstics of a number of dimension reduction implementations.
Umap Fast In R.