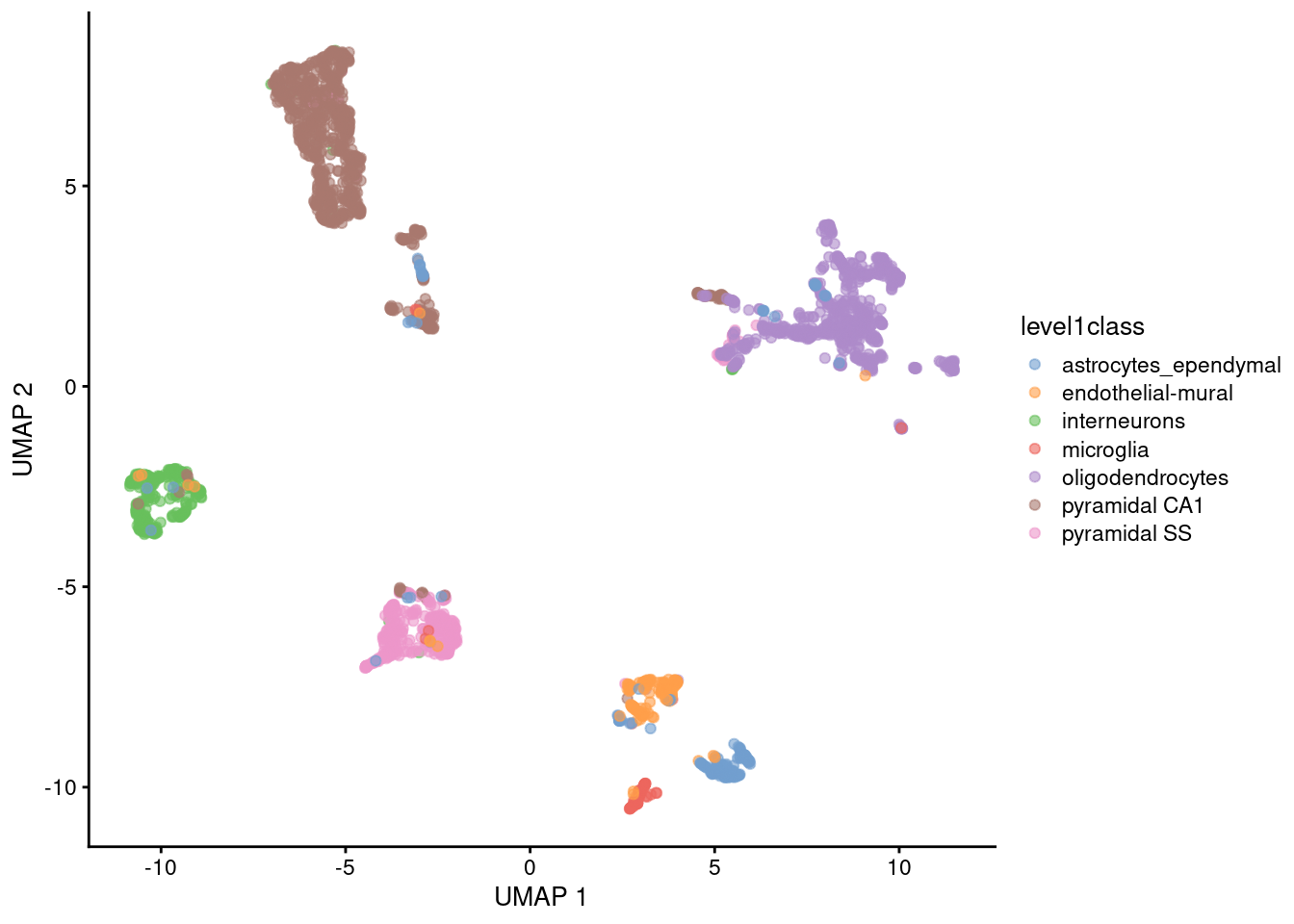
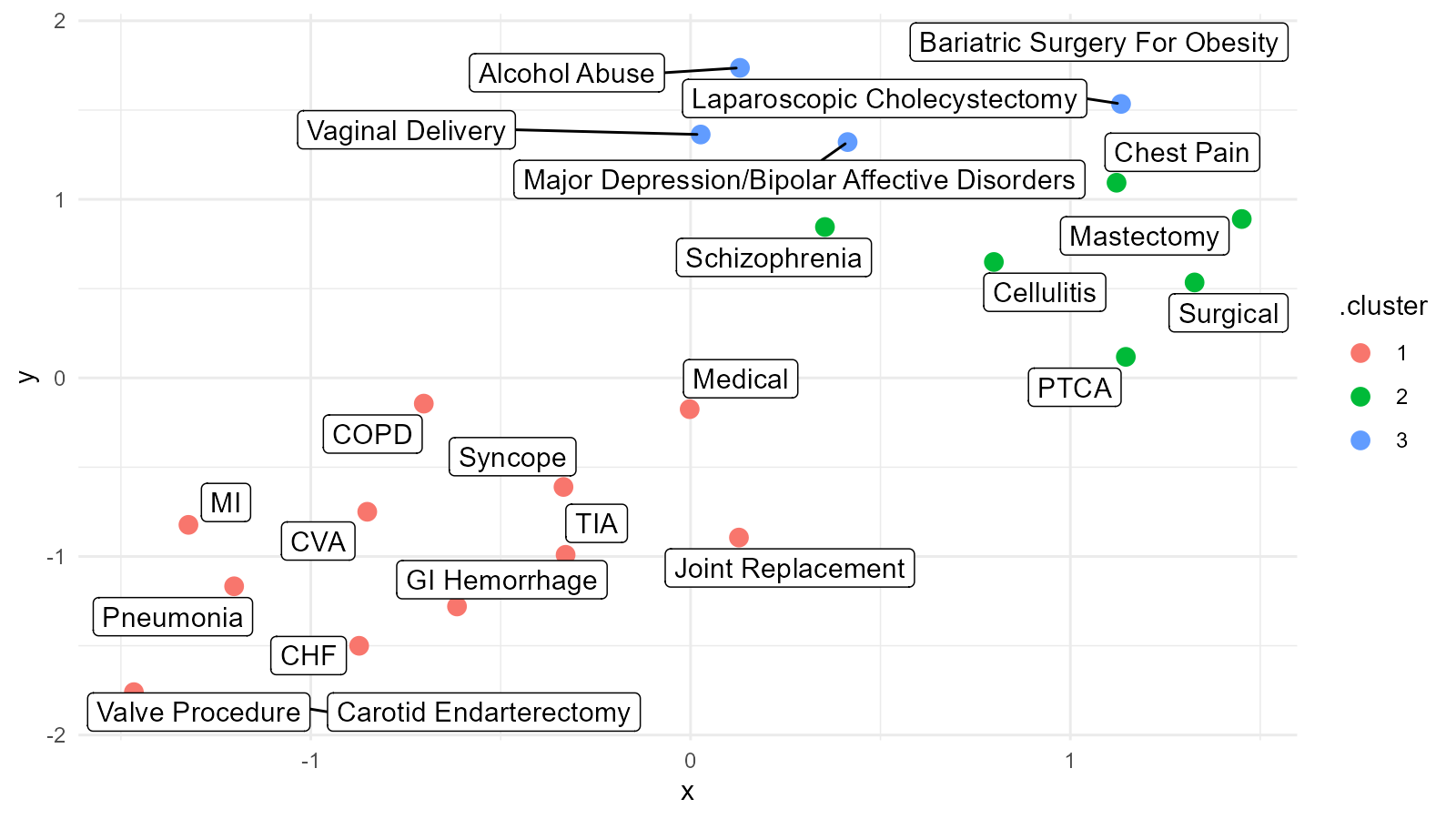
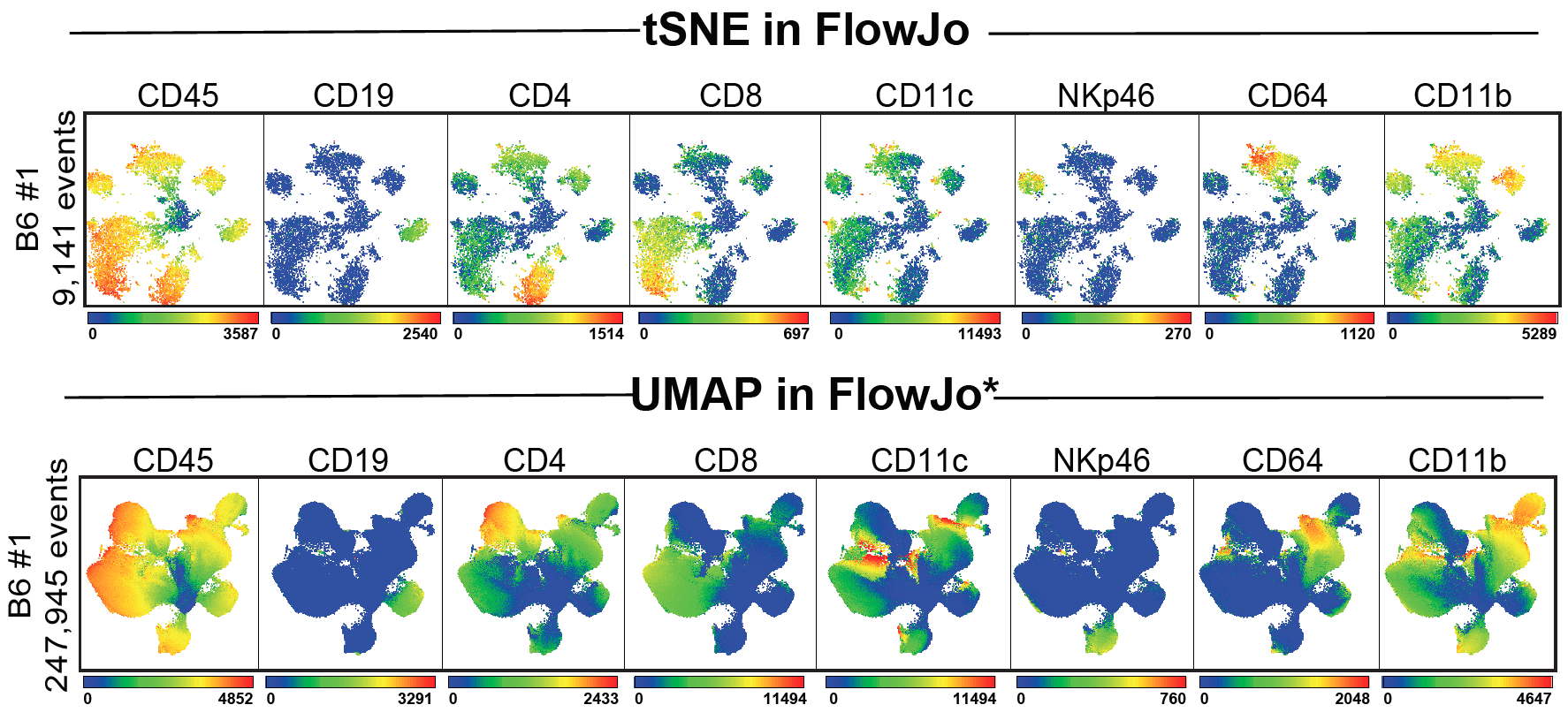
Umap Basics. Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. It seeks to learn the manifold structure of your data and find a low dimensional embedding that preserves the essential topological structure of that manifold. It is designed to be compatible with scikit-learn, making use of the same API and able to be added to sklearn pipelines. The algorithm is founded on three assumptions about the data. UMAP is a fairly flexible non-linear dimension reduction algorithm. UMAP is a general purpose manifold learning and dimension reduction algorithm. Dimensionality reduction is a powerful tool for machine learning practitioners to visualize and understand large, high dimensional datasets. In other words, this gives a locally adaptive exponential kernel for each data point, so the distance metric varies from point to point.

Umap Basics. It is designed to be compatible with\nscikit-learn, making use\nof the same API and able to be added to sklearn pipelines. Alex Diaz-Papkovich, Luke Anderson-Trocmé & Simon Gravel. The algorithm is founded on three assumptions about the data. The data is uniformly distributed on Riemannian manifold; UMAP is a dimensionality reduction algorithm and a powerful data analysis tool. If you are\nalready familiar with sklearn you should be able to use UMAP as a drop\nin replacement for t-SNE and other dimension reduction classes. Umap Basics.
If you are\nalready familiar with sklearn you should be able to use UMAP as a drop\nin replacement for t-SNE and other dimension reduction classes.
Good old PCA on the other hand is deterministic and easily understandable with basic knowledge of linear algebra (matrix multiplication and eigenproblems), but is just a linear reduction in contrast to the non-linear reductions of t-SNE and UMAP.
Umap Basics. Here ρ is an important parameter that represents the distance from each i-th data point to its first nearest neighbor. The algorithm is founded on three assumptions about the data. The\nresulting class is quite flexible, but here we will walk through simple\nusage on some basic (and somewhat contrived) data just to demonstrate\nhow to get it running on data. \n import numpy as np \n import sklearn. datasets \n import umap \n import umap . plot \n import umap . utils as utils \n import umap . aligned_umap \n import . Alex Diaz-Papkovich, Luke Anderson-Trocmé & Simon Gravel. To make use of UMAP as a data transformer we first need to fir the model with the training data.
Umap Basics.