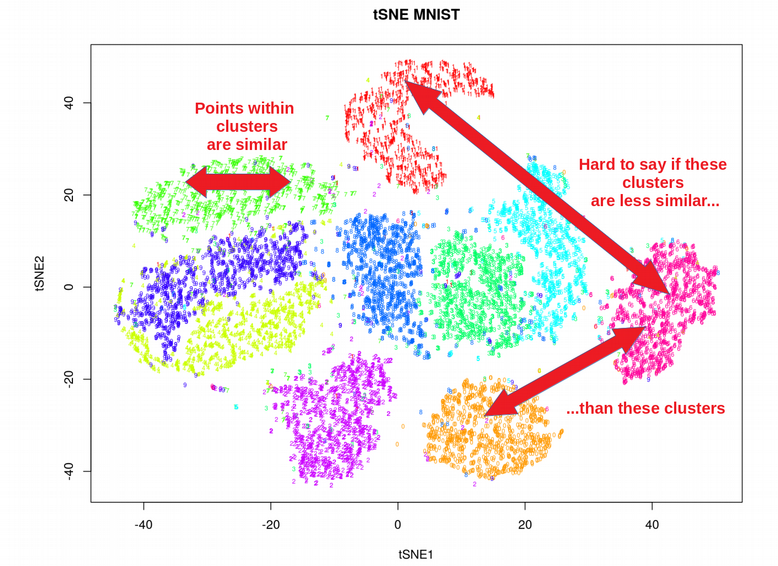
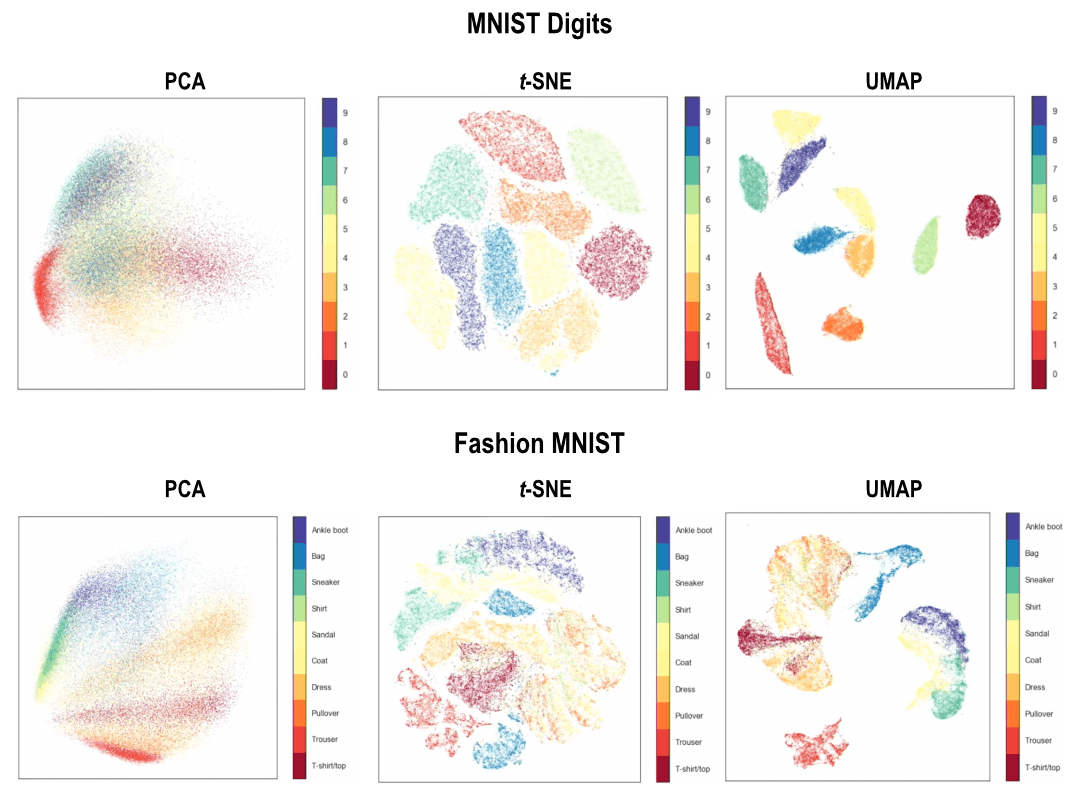
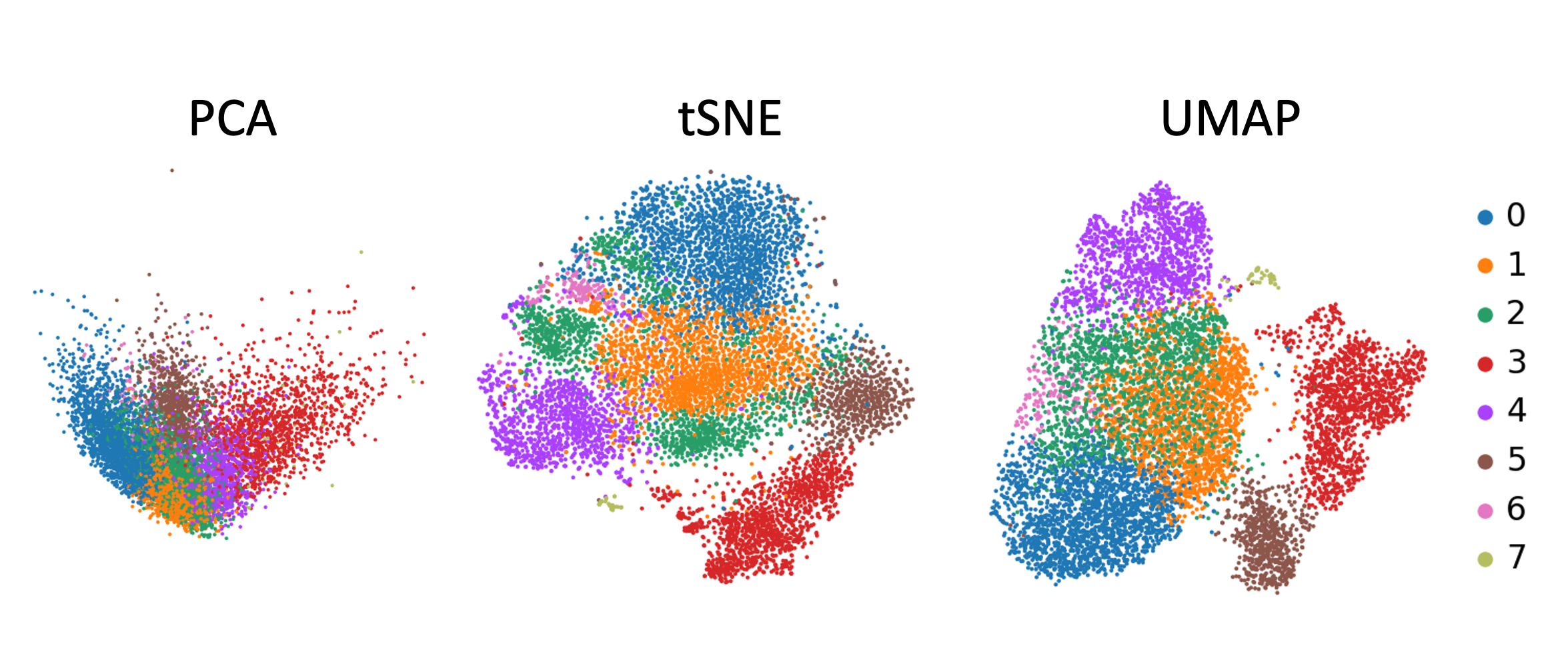
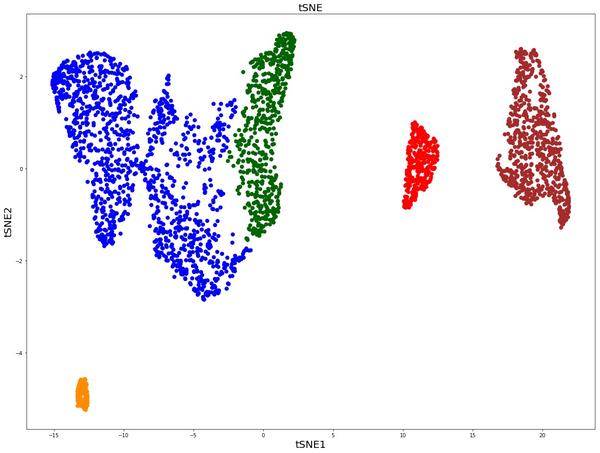
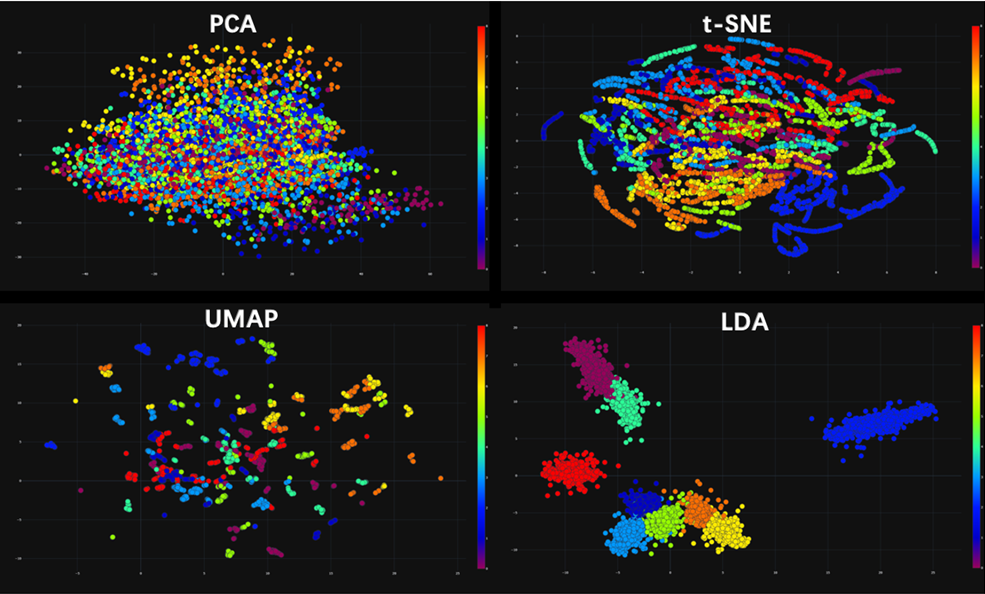
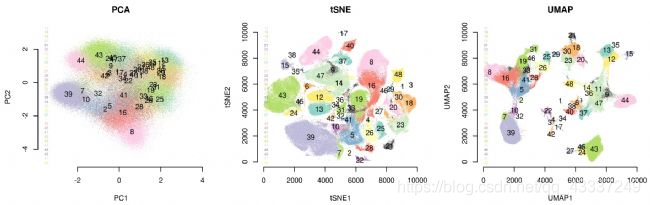
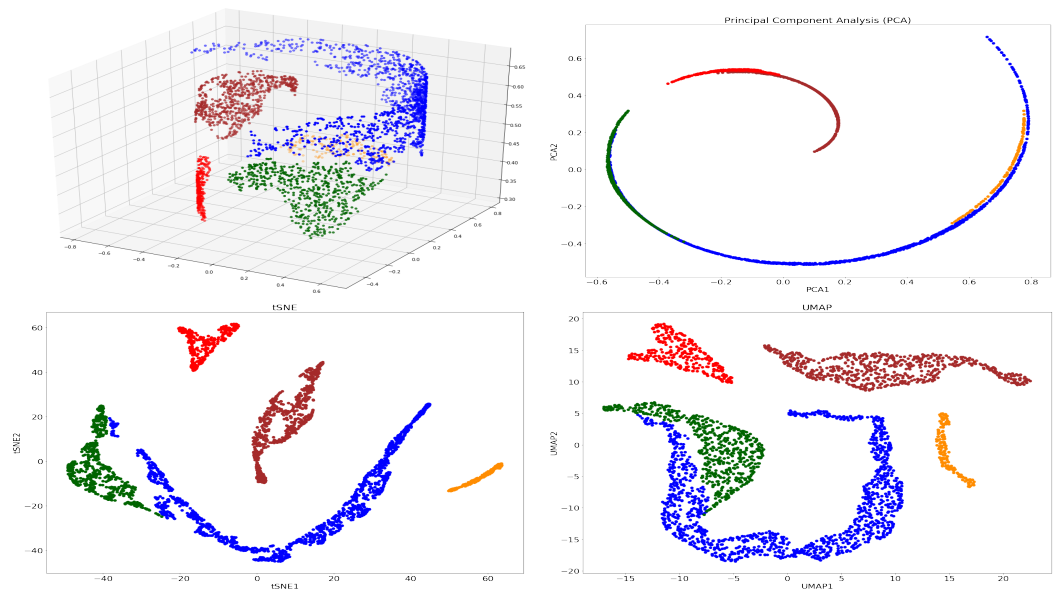
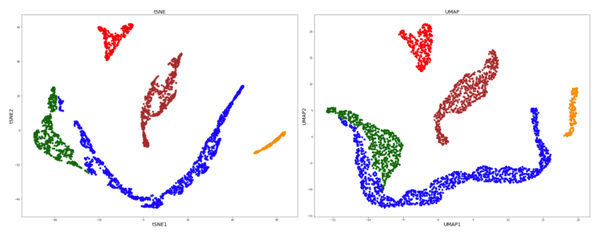
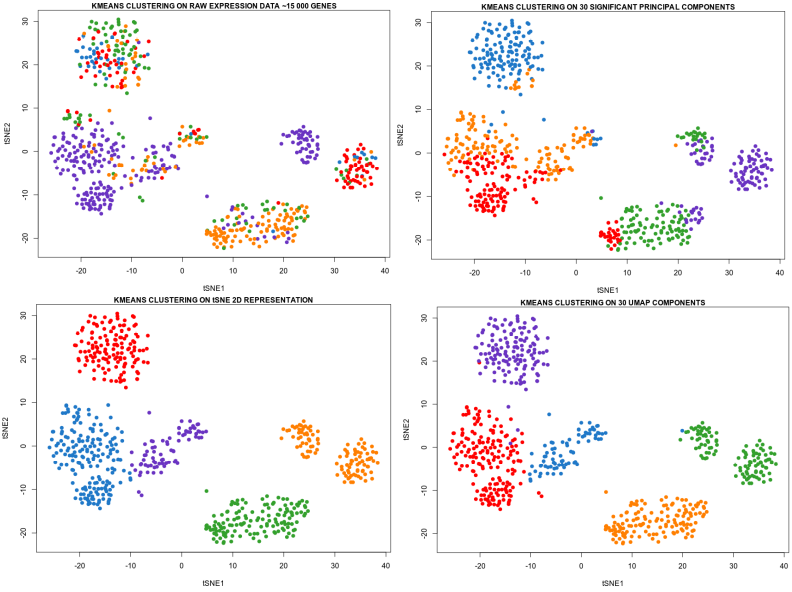
Umap And Tsne. Despite their popularity, however, little work has been done to study their full span of differences. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. Noise-contrastive estimation can be used to optimize t -SNE, while UMAP relies on negative sampling, another. tSNE and UMAP are popular dimensionality reduction algorithms due to their speed and interpretable low-dimensional embeddings. In the example below, we see how easy it is to use UMAP as a drop-in replacement for scikit-learn's manifold. My studies recently started to involve high dimensional data analysis. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. Common data analysis pipelines include a dimensionality reduction step for visualising the data in two dimensions, most frequently performed using t-distributed stochastic neighbour embedding. In this work, we uncover their conceptual connection via a new insight into contrastive learning methods.
Umap And Tsne. Our approach automatically learns the relative contribution of each modality to a concise representation of cellular identity that promotes discriminative features but suppresses noise. My studies recently started to involve high dimensional data analysis. Get the latest on events, weather, sports, entertainment, and more. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. Common data analysis pipelines include a dimensionality reduction step for visualising the data in two dimensions, most frequently performed using t-distributed stochastic neighbour embedding. Umap And Tsne.
My studies recently started to involve high dimensional data analysis.
It is now read-only. ropensci-archive / umapr.
Umap And Tsne. Users' confusion often revolve around following questions: UMAP vs t-SNE, which is better? The cloud of confusion between UMAP vs t-SNE has been going for a while. t-SNE and UMAP seem similar in their principles, but the outcomes are (sometimes) dramatically different. On eight datasets, j-SNE and j-UMAP produce unified embeddings that better agree with known cell types and that harmonize RNA and protein velocity landscapes. In this work, we uncover their conceptual connection via a new insight into contrastive learning methods. In the example below, we see how easy it is to use UMAP in R.
Umap And Tsne.