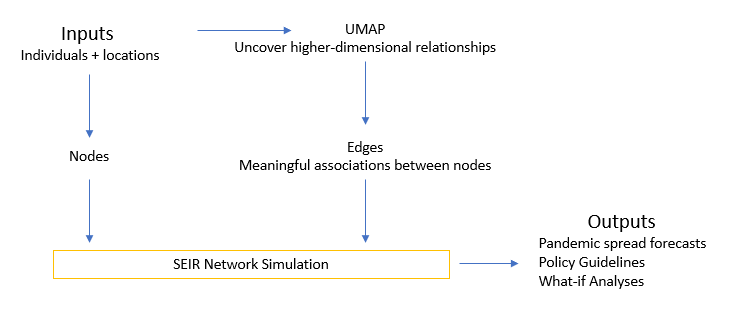
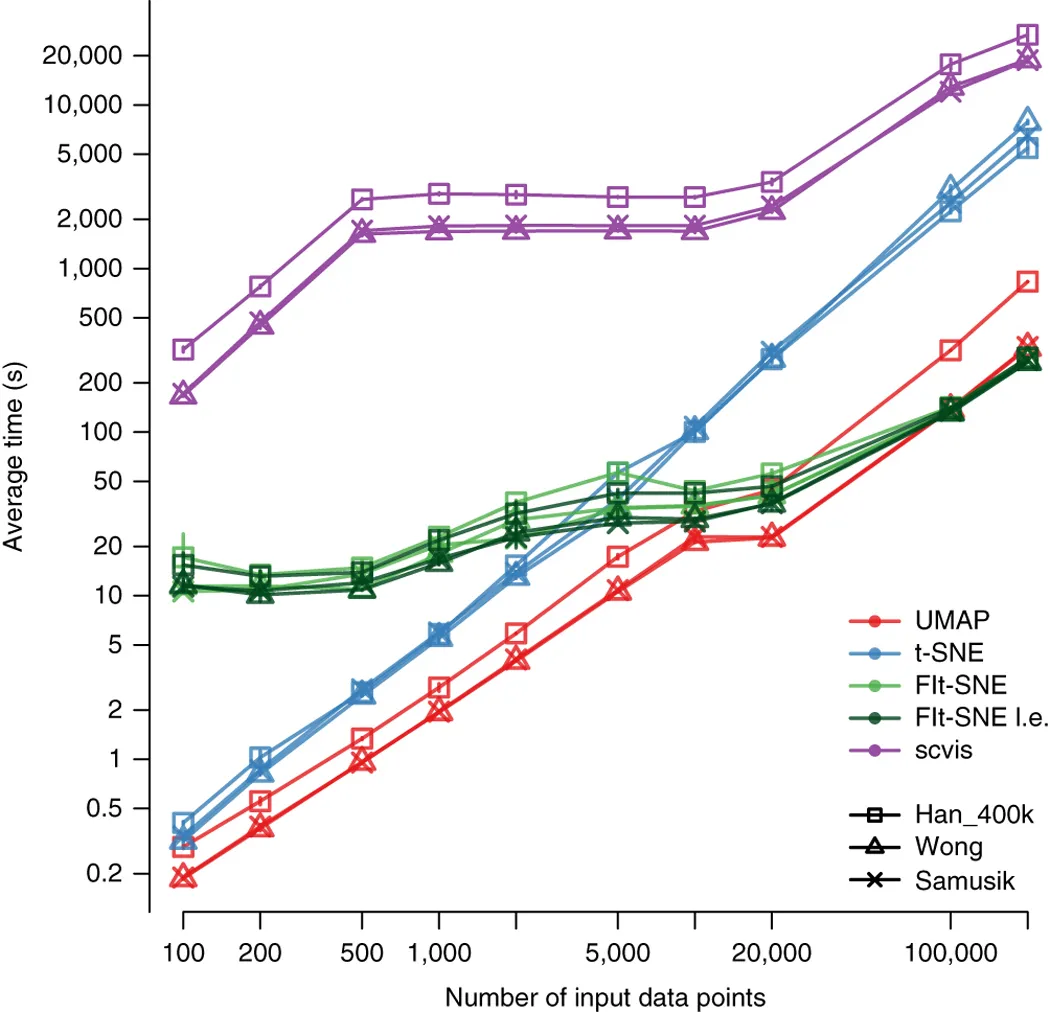
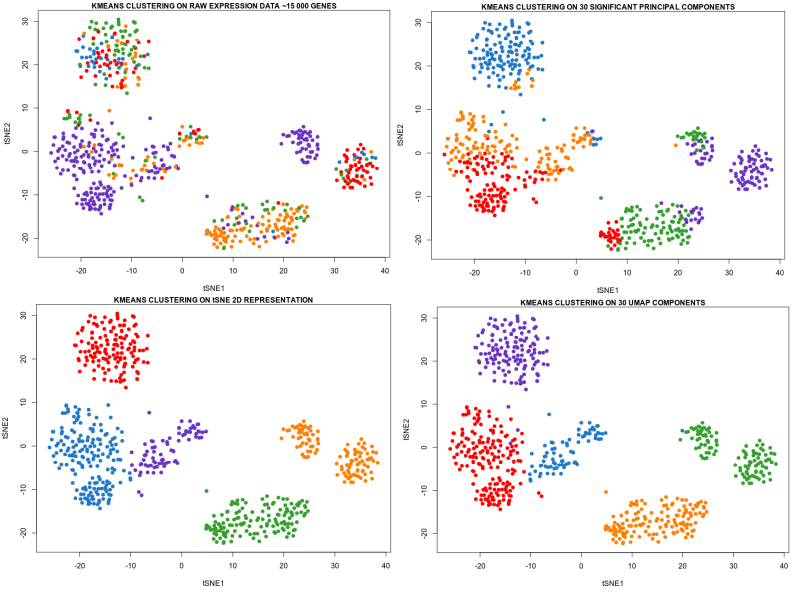
Umap Analyse. It has become common in visualizing the ancestral. Uniform Manifold Approximation and Projection (or UMAP) is a new dimension reduction technique that can be used to visualize patterns of clustering in high-dimensional data. The R package umap described in this vignette is a separate work that provides two implementations for using UMAP within the R environment. Uniform Manifold Approximation and Projection (UMAP) is an algorithm for dimensional reduction. Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. The comparison of the raw high-dimensional data with the projections obtained using different dimensionality reduction methods based on various metrics showed that UMAP has superior performance when compared with linear reduction methods (PCA and tICA) and has competitive performance and scalable computational cost. UMAP groups deletion mutants with shared protein function We assigned groups, or clusters, to deletion mutants with similar transcriptional responses using the Louvain community detection. It is similar to PCA (Principal Component Analysis) in terms of speed and resembles tSNE to reduce dimensionality while preserving as much information of the dataset as possible.

Umap Analyse. In general, a common practice is to validate UMAP's convergence based on a downstream task. UMAP is a dimensionality reduction algorithm and a powerful data analysis tool. Below is a selection of uses cases of UMAP being used for interesting explorations of intriguing. Uniform Manifold Approximation and Projection (UMAP) is an algorithm for dimensional reduction. It has become common in visualizing the ancestral. Umap Analyse.
The dimensions of matrices (the number of rows/cases by the number of columns/species) differed between studies, allowing these.
The comparison of the raw high-dimensional data with the projections obtained using different dimensionality reduction methods based on various metrics showed that UMAP has superior performance when compared with linear reduction methods (PCA and tICA) and has competitive performance and scalable computational cost.
Umap Analyse. Uniform Manifold Approximation and Projection (or UMAP) is a new dimension reduction technique that can be used to visualize patterns of clustering in high-dimensional data. What pitfalls should we consider when interpreting UMAP or t-SNE plot for single-cell RNA-Seq? Today we are going to dive into an exciting dimension reduction technique called UMAP that dominates the Single Cell Genomics nowadays. Basic UMAP Parameters ¶ UMAP is a fairly flexible non-linear dimension reduction algorithm. In general, a common practice is to validate UMAP's convergence based on a downstream task.
Umap Analyse.