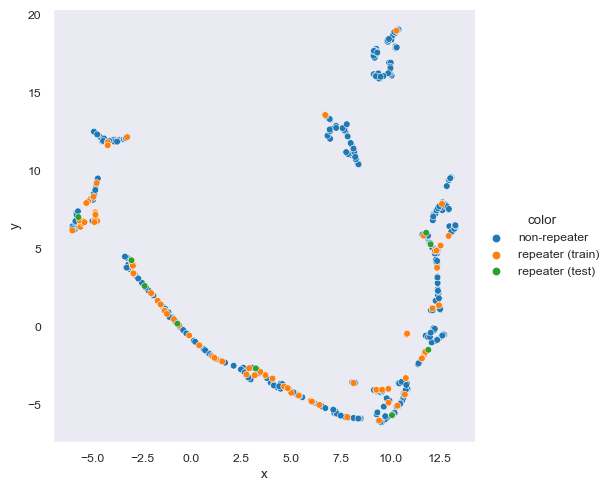
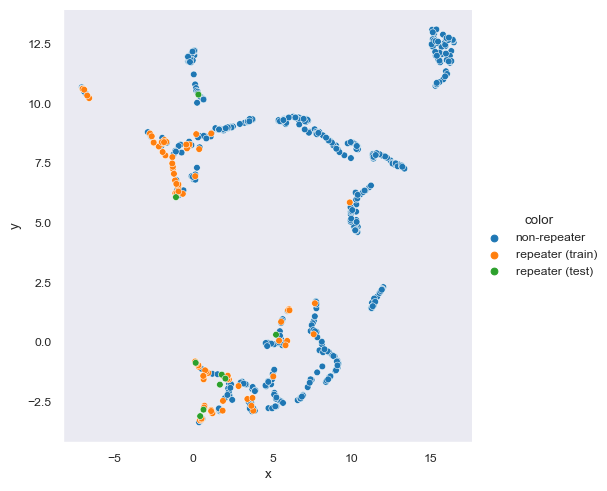
Umap And Hdbscan. HDBSCAN does work well with UMAP, with the caveat that with UMAP's uniform density assumption it too does not preserve density well. Data exploration with UMAP / hdbscan. Better results are found for these new approaches. With the embeddings ready, reduce the dimensionality down to five features using cuML's UMAP. The chatintents package makes it easy to implement this tuning process. The performances of UMAP and HDBSCAN are systematically compared to the other clustering analysis approaches used in EELS in the literature using a known synthetic dataset. This is somewhat controversial, and should be attempted with care. Furthermore, UMAP and HDBSCAN are showcased in a real experimental dataset from a core-shell nanoparticle of iron and manganese oxides, as well as the triple combination nonnegative matrix factorization-UMAP-HDBSCAN.

Umap And Hdbscan. For the sake of brevity, the results of the feature correlation. With the embeddings ready, reduce the dimensionality down to five features using cuML's UMAP. It extends DBSCAN by converting it into a hierarchical clustering algorithm, and then using a technique to extract a flat clustering based in the stability of clusters. In our In the Koumura dataset , HDBSCAN on the PCA projections gives the closest match to human clustering, where homogeneity is higher with HDBSCAN/UMAP and completeness is higher with HDBSCAN/PCA. HDBSCAN does work well with UMAP, with the caveat that with UMAP's uniform density assumption it too does not preserve density well. Umap And Hdbscan.
UMAP can be used as an effective preprocessing step to boost the performance of density based clustering.
HDBSCAN does work well with UMAP, with the caveat that with UMAP's uniform density assumption it too does not preserve density well.
Umap And Hdbscan. This implies that the bermude area is much more heterogeneous than a single. The chatintents package makes it easy to implement this tuning process. Furthermore, UMAP and HDBSCAN are showcased in a real experimental dataset from a core-shell nanoparticle of iron and. Many Git commands accept both tag and branch names, so creating this branch may cause unexpected behavior. In our In the Koumura dataset , HDBSCAN on the PCA projections gives the closest match to human clustering, where homogeneity is higher with HDBSCAN/UMAP and completeness is higher with HDBSCAN/PCA.
Umap And Hdbscan.