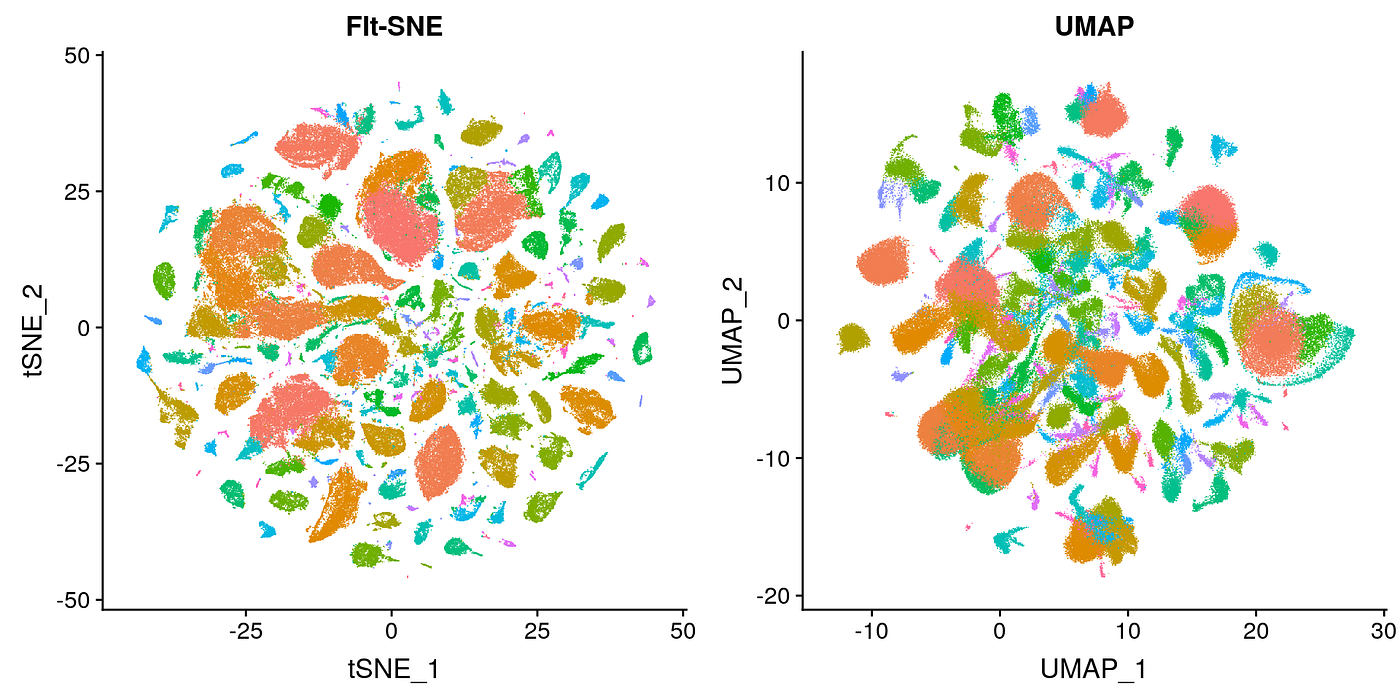
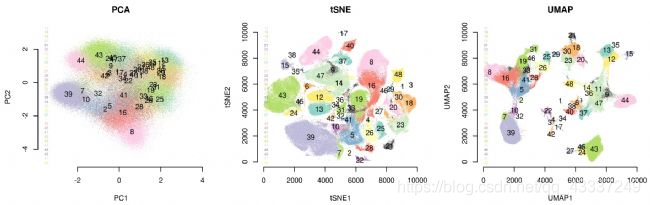
Umap And T Sne. In this work, we uncover their conceptual connection via a new insight into contrastive learning methods. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. In this story, we are gonna go through three Dimensionality reduction techniques specifically used for Data Visualization: PCA, t-SNE, LDA and UMAP. What pitfalls should we consider when interpreting UMAP or t-SNE plot for single-cell RNA-Seq? Both algorithms minimize their loss functions by. The fundamental reasons for this and, more generally, the exact relationship between t -SNE and UMAP have remained unclear. At the core of both t-SNE and UMAP are loss functions that make similar points attract each other and push dissimilar points away from each other. The latter category, which includes SNE (Stochastic Neighbor Embedding), t-SNE (t-distributed Stochastic Neighbor Embedding), and UMAP (Uniform Manifold Approximation and Projection), will be the focus of this discussion.
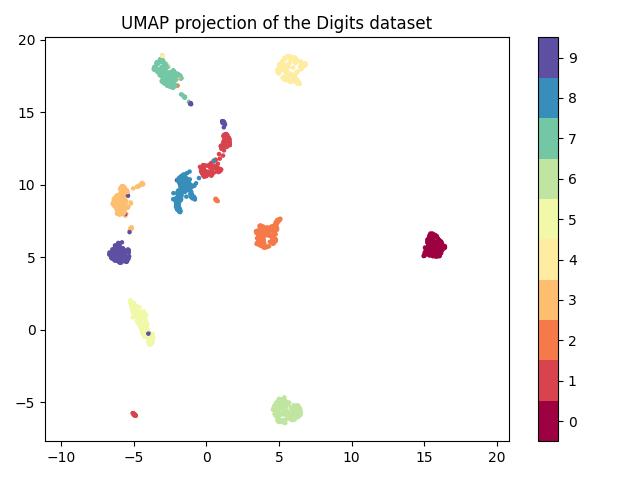
Umap And T Sne. In this article, we'll give you the answers in the simplest manner possible. Indeed, both tSNE and UMAP were designed to predominantly preserve local structure that is to group neighboring data points together which indeed provides a very informative visualization of heterogeneity in your data. We are going to explore them in details using the Sign Language MNIST Dataset, without going in-depth with the maths behind the algorithms. In this work, we uncover their conceptual connection via a new insight into contrastive learning methods. Noise-contrastive estimation can be used to optimize t -SNE, while UMAP relies on negative sampling, another. Umap And T Sne.
At the core of both t-SNE and UMAP are loss functions that make similar points attract each other and push dissimilar points away from each other.
The latter category, which includes SNE (Stochastic Neighbor Embedding), t-SNE (t-distributed Stochastic Neighbor Embedding), and UMAP (Uniform Manifold Approximation and Projection), will be the focus of this discussion.
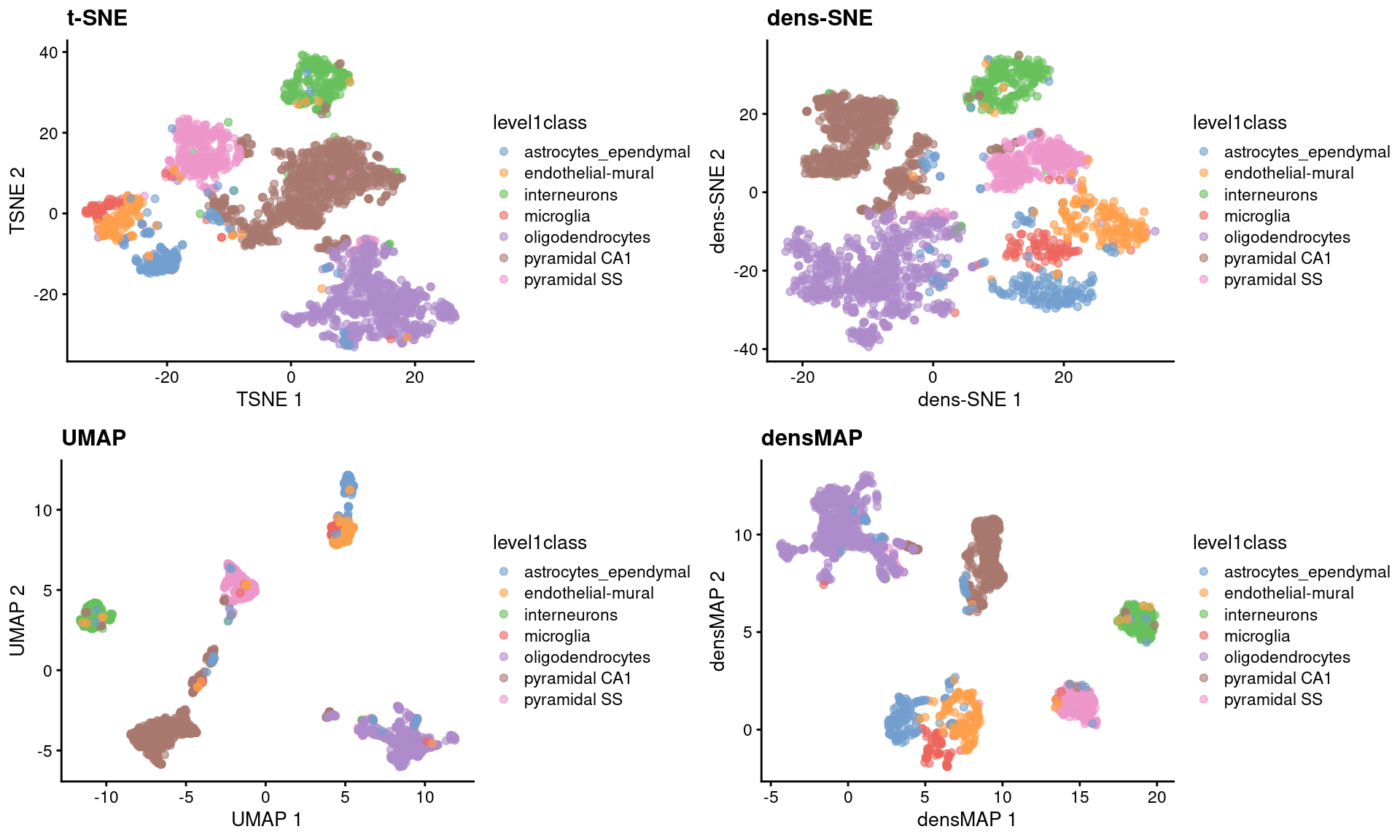
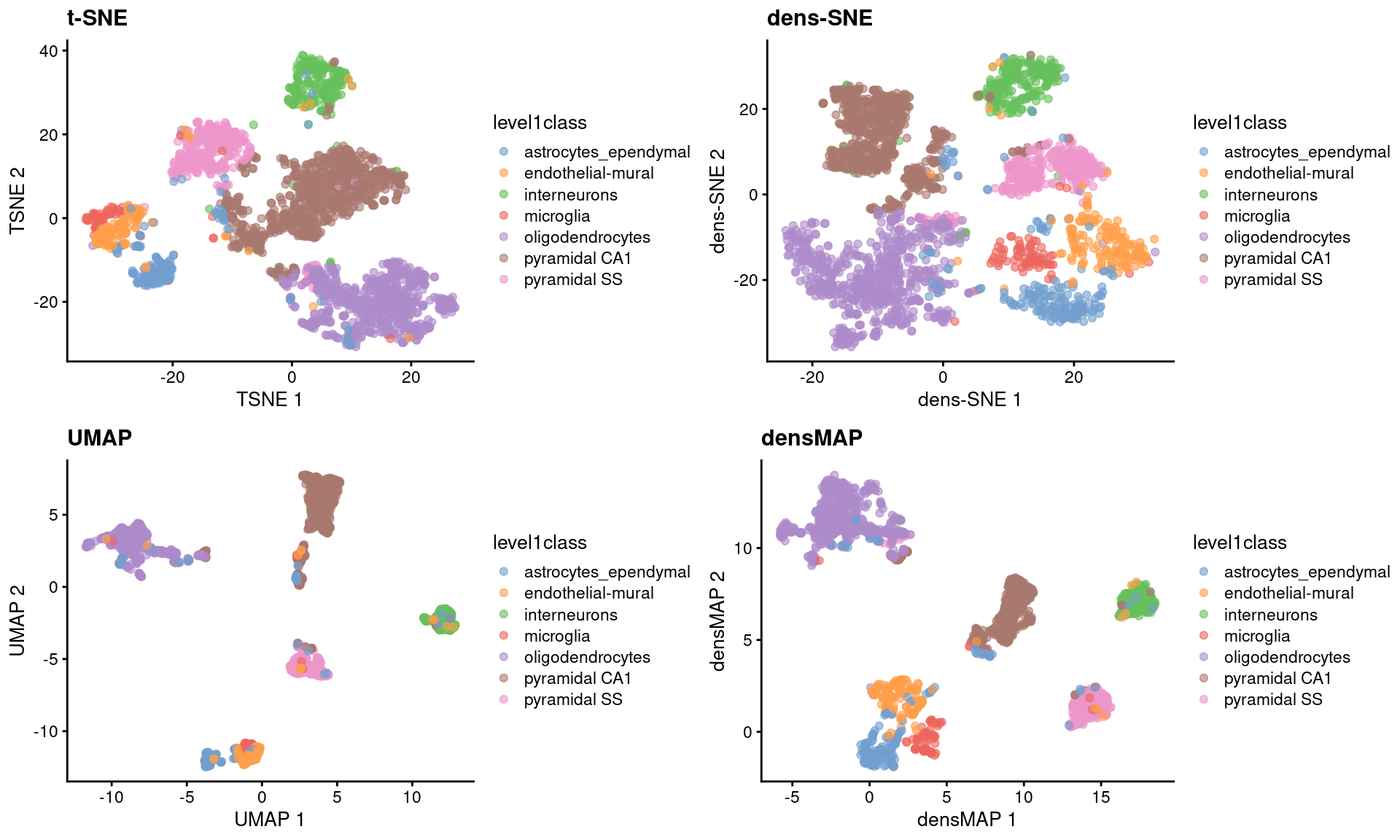
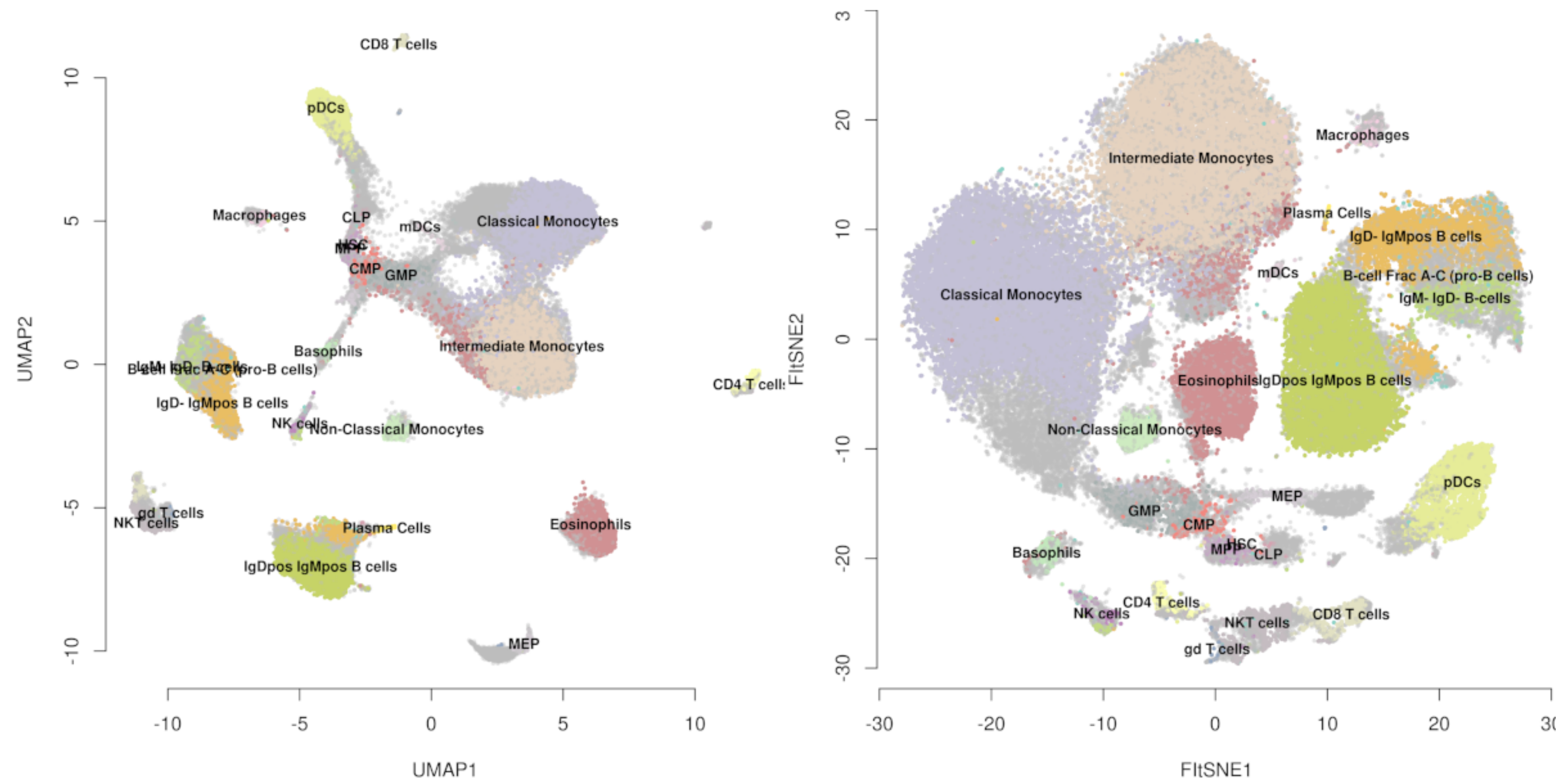
Umap And T Sne. We are going to explore them in details using the Sign Language MNIST Dataset, without going in-depth with the maths behind the algorithms. As the number of data points increase, UMAP becomes more time efficient compared to TSNE. However, one limitation with UMAP and t-SNE is that they can only capture the local structure of the data, the global structure of the data is not faithfully preserved. Noise-contrastive estimation can be used to optimize t -SNE, while UMAP relies on negative sampling, another. UMAP and t-SNE were ran independently on the dataset to qualitatively compare the distribution of clustered cell types in both reduced dimensions.
Umap And T Sne.